Lapidus Group
Our group studies protein folding, dynamics and aggregation using laser spectroscopy and microfluidics. We are currently interested in proteins involved in neurodegenerative diseases and protein dynamics in cell-like conditions.
Lab Publications
current Projects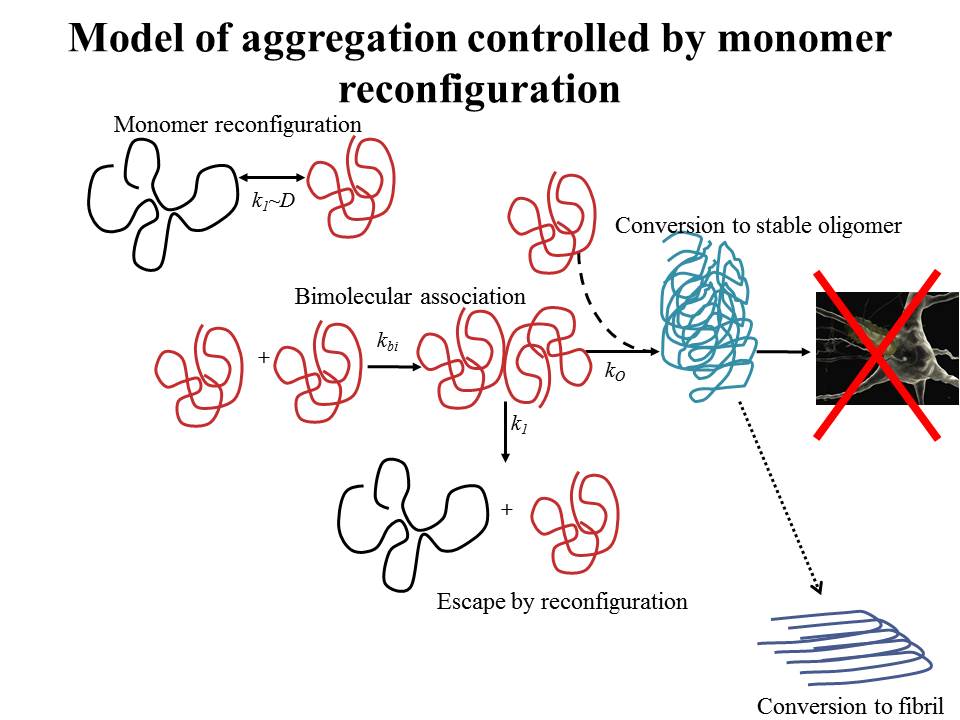
Protein aggregation is phenomena that underlies many diseases, such as Alzheimer's, Parkinson's and Type II Diabetes. The aggregated proteins eventually form long fibrillar structures but usually originate from disordered structures and small disordered oligomers are likely the toxic species that causes cell death. We hypothesize that the formation of the first oligomer is controlled by the speed of monomer reconfiguration, intramolecular diffusion. When diffusion is fast, monomers can escape by reconfiguring to a less "sticky" conformation, but as diffusion slows, aggregation becomes more likely. Using measurements of Trp-Cys quenching, we have demonstrated this model in a variety of disease-associated proteins, including α-synuclein, the Alzheimer's peptide and mammalian prion proteins.
Phase separation of Protein and RNA solutions
There have been many observations of spontaneous formation of liquid-like condensates
in both the test tube and the cell, but these cases are usually for disordered proteins
and RNAs. We have shown that condensation is also possible for folded proteins and
RNAs. Coarse grained models can predict the phenomenon based on the net charge and
size of the molecules.
Protein-Protein interactions in cell conditions
In a test tube, the concentration of proteins are usually low so we can treat the molecules like billiard balls that occasional collide. But in a cell, the concentration of proteins is very high so that transient interactions are significant. We use Trp-Cys quenching to observe bimolecular interactions and see distinct differences for different sequences. All atom MD simulations (in collaboration with Michael Feig) show good agreement with experiments.
Techniques
We use a variety of optical methods to observe biological molecules, including fluorescence, absorption and circular dichroism. Optical techniques can look at dynamics on very fast timescales, but the process needs to be prompted rapidly as well.
Trp-Cys Quenching
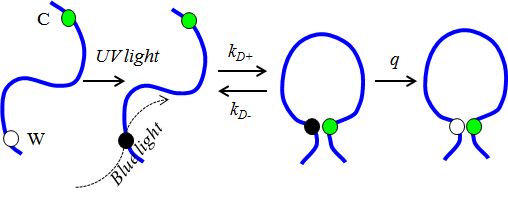
We use a specialized technique to observe dynamics of disordered or unfolded proteins - Trp-Cys contact quenching. Tryptophan (W) is excited to a long-lived triplet state that is quenched on contact with cysteine (C). The rates we measure give us an estimate of the volume of the chain and the rate of reconfiguration.
Microfluidic Mixing
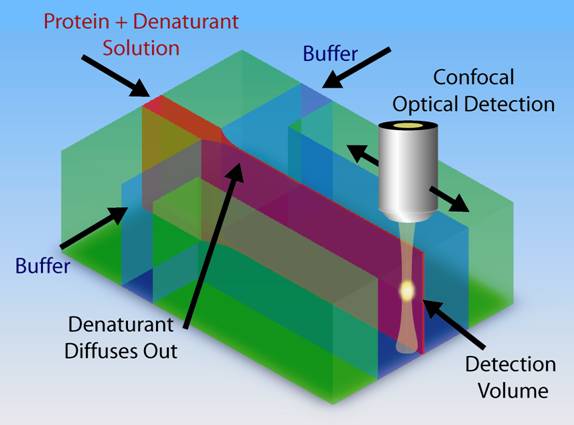
We also use microfluidic mixers which can mix two solution in as little as a few microseconds. This mixer can be used to observe dynamics with intrinsic Trp fluorescence, Fluorescence Resonance Energy Transfer (FRET) or Fast Photochemical Oxidation of Proteins (FPOP).
Lab Members
Lisa Lapidus got her PhD at Harvar d University in Atomic Physics studying electrons in a Penning trap (this video with Jim Carrey and Conan O'Brien is based on that research). For her postdoctoral
work, she switched fields to biophysics, studying protein folding with Bill Eaton and RNA folding with Steve Chu. Lisa moved to MSU in 2004.
d University in Atomic Physics studying electrons in a Penning trap (this video with Jim Carrey and Conan O'Brien is based on that research). For her postdoctoral
work, she switched fields to biophysics, studying protein folding with Bill Eaton and RNA folding with Steve Chu. Lisa moved to MSU in 2004.
We are currently looking for new students!